Lab Research People Publications Facebook
Research in our lab is aimed at studying root-microbe interactions in leguminous plant species. Specifically, we examine the role of miRNAs and plant hormones on legume nodule development and the role of plant rhizodeposits and management practices on plant-associated microbial communities.
Research Projects:
Plants are colonized by a number of microbes in native environments. Through a collaboration with Heike Bucking's lab, we seek to understand what mechanisms plant choose to allow colonization by specific species or strains of microbes depending on their nutritional needs. For example, when colonized simultaneously by both rhizobia and arbuscular mycorrhizal fungi, do the plans reward one symbiont more than the other depending on the nutritional or other benefits the symbiont offers. This work is funded by a grant award from NSF-USDA's Plant Biotic Interactions program.
We are also evaluating if and how plants select and reward specific species of rhizobia based on their nitrogen fixation capacity. We are collaborating with interdisciplinary groups to not only gain a better understanding of this process, but also evaluate potentially transformative approaches to modify root surfaces for enhanced colonization by desirable bacteria. This work is funded by a grant award from NSF-EPSCoR. Collaborators include Zhengrong Gu, Volker Brozel, Heike Bucking and Nicholas Butzin from SDSU, Ramana Gadhamshetty and Rajesh Sani from SD Mines, and Carol Lushbough and Etienne Gnimpieba from USD.
We identified that enhanced auxin signaling inhibits nodule formation in soybean by suppressing cytokinin action (Turner et al. 2013 Plant Physiology). Subsequent experiments using auxin associated miRNAs indicated that they might play distinct roles in determinate vs indeterminate nodule formation. (Mao et al. 2013 Plant Signaling and Behavior, Bustos-Sanmamed et al. 2013 Functional Plant Biology). Subsequent experiments revealed that miR160 dictates developmental stage-specific auxin-cytokinin balance during soybean nodule development (Nizampatnam et al., 2015 Plant J). This work was funded by a grant award from USDA-NIFA.
We identified 120 previously unknown miRNA genes from soybean including multiple novel miRNA families. In the soybean genome, genes encoding miRNAs are primarily intergenic and a small percentage were intragenic or less than 1000 bp from a protein-coding gene, suggesting potential co-regulation between the miRNA and its parent gene. Difference in number and orientation of tandemly duplicated miRNA genes between orthologous genomic loci indicated continuous evolution and diversification (Turner et al 2012 BMC Genomics). We determined that a set of novel soybean/legume-specific miRNAs that target resistance genes and other components of plant defense mechanisms play a key role in regulating nodule formation in soybean (Li et al 2010 Plant Physiology). This work was funded by a grant award from USDA-NIFA.
We have enhanced current assay methods for miRNA abundance and activity. We determined that Hairpin Priming Is Better Suited than In Vitro Polyadenylation to Generate cDNA for Plant miRNA qPCR [Adhikari et al. 2013 Mol Plant]. We optimized hairpin/stem-loop qPCR assays through multiplexed cDNA synthesis of U6 and miRNAs and demonstrated the use of miR1515 as a house-keeping miRNA in soybean [Turner et al. 2013 Plant Signaling and Behavior.]. We enhanced miRNA activity assay using a qPCR method named 'quantitative Amplification of Cleaved Ends or qACE' [Damodaran et al. under review]. This work was funded by a grant award from USDA-NIFA.
We have also identified additional mechanisms by which auxin levels might be regulated during soybean nodule development. For example, regulation of IAA-aminoacid conjugating GH3 enzymes that determine free auxin levels [Damodaran et al. 2017 IJMS], and post-translational regulation of HD-ZIPIII proteins that regulate nodule vascular development appear to be crucial for proper nodule development [Damodaran et al. 2019 IJMS]. Additional genes and mechanisms that dictate auxin levels and/or sensitivity are being evaluated using a variety of approaches. This work was funded by a grant awards from NSF-IOS and USDA-NIFA.
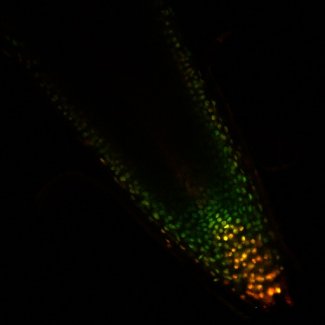
Soybean nodules are comprised of two major developmental zones, (i) the nodule primordium (Npr) in the middle, and (ii) the surrounding nodule parenchyma (Npa), differentiate from root cortex cells. The Npr ultimately gives rise to the N-fixation zone; the Npa houses vascular bundles and structural components of the nodule. It is not known what early signaling pathways cause distinct development of these nodule zones. Neither is it known what mechanisms regulate distinct gene expression profiles in Npr and Npa cell types. We use functional genomics and imaging approaches to determine developmental and molecular processes that occur during the development of these two nodule zones
We developed quantitative imaging methods in collaboration with Steve Smith's lab at SDSMT to concurrently image auxin and cytokinin transcriptional outputs at cell level in roots and nodules. Our results indicated that distinct auxin-cytokinin output ratios might dictate the development of distinct cell types in nodules [Fisher et al. 2018 Plant Cell and Environment].
This work has received very high Attention Score compared to outputs of the same age and source (96th percentile). This work was funded by a grant awards from NSF-IOS and NSF-EPSCoR.
High bacterial density and diversity near plant roots has been attributed to rhizodeposit compounds that serve as both energy sources and signal molecules. To determine the role of rhizodeposit flavonoids in plant-microbe interactions in the rhizosphere, we silenced the biosynthesis of isoflavonoids, a major component of soybean rhizodeposits, using RNA interference in hairy root composite plants, and examined changes in rhizosphere bacteriome diversity. Our results suggested a temporal gradient effect of rhizodeposit isoflavonoids on the rhizosphere and that the hairy root transformation process itself significantly altered rhizosphere bacterial diversity [White et al. 2014 MPMI accepted]. The detailed method was published as an invited article [White et al. 2015 Bio-Protocol]. We subsequently used next-generation sequencing to determine that isoflavonoids had a small effect on bacterial community structure, and in particular on the abundance of Xanthomonads and Comamonads; and that hairy roots and untransformed plant roots had ~74% similarity in colonization trend of rhizosphere bacteria [White et al. 2017 Environment Microbiology]. This work was funded by a grant awards from USDA-NIFA (SD-AES).
We determined that nitrogen fixed by kuraclover can replace nitrogen fertilization in about three years after planting [phys.org feature]. We are continuing to explore the impact of management practices and other factors e.g. landscape position and soil temperature on microbial community composition and activity [Sekaran et al. 2019 GCB Bioenergy]. This work was funded by a grant awards from DOE and USDA-NIFA.
Research Funding:
- Research in the lab supported by past and/or present funding from
- South Dakota Soybean Research and Promotion Council
- USDA - NIFA
- DOE - Sun Grant Initiative
- NSF-EPSCoR (BioSNTR, 2DBEST)
- NSF PGRP (CAREER award)





